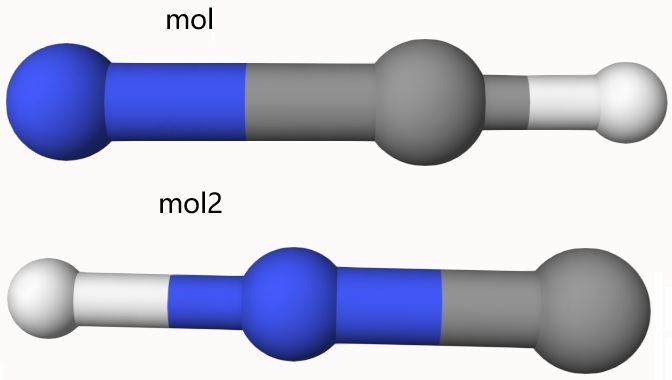
mol and mol2
The keyword mol provides the initial molecular coordinates.
For certain tasks, such as transition state searches, a second structure must be specified using the mol2 option.
Explicit XYZ Coordinates
You can simply provide XYZ coordinates in Ångström:
mol
O 0.00000000000000 0.05011194954430 0.05011194954224
H 0.00000000000000 -0.06080277603381 1.01069082652926
H 0.00000000000000 1.01069082648951 -0.06080277607149
endXYZ File Name
You can provide the name of the XYZ file (including the path, if necessary):
PDB File Name
You can provide the name of the PDB file (including the path, if necessary):
Theoretical Background
XYZ File Format
-
An XYZ file is a text format used to describe the geometry of a molecule. Its structure is as follows:
- Number of atoms: The first line contains an integer representing the total number of atoms in the molecule.
- Comment line: The second line is typically a comment, which may provide additional information about the molecule.
- Atomic coordinates: Each subsequent line includes the element symbol of an atom followed by its Cartesian coordinates (x, y, z), typically expressed in Ångströms.
3
water molecule
O 0.000000 0.000000 0.116863
H 0.000000 0.764949 -0.467453
H 0.000000 -0.764949 -0.467453PDB File Format
The Protein Data Bank (PDB) file format is a comprehensive text format widely used to store three-dimensional structural information of biological macromolecules, such as proteins and nucleic acids.
Each PDB file contains detailed information, including the molecule's name, source species, sequence, secondary structure, and crystallographic parameters.
In Qbics, only atomic records are used—specifically, lines beginning with
ATOM and HETATM.
-
Each atomic record in a PDB file provides detailed information about an atom:
-
Record Type: Indicates the type of record, such as
ATOMorHETATM. - Atom Serial Number: A unique identifier for each atom.
- Atom Name: The element symbol, possibly with additional characters to distinguish atoms within the same residue.
-
Residue Name: The name of the residue (e.g.,
ALAfor alanine). - Residue Sequence Number: The position of the residue in the sequence.
- X, Y, Z Coordinates: The atom’s Cartesian coordinates in Ångströms.
- Occupancy: The fraction of the molecule present at this position (a crystallographic parameter not used by Qbics).
- Temperature Factor (B-factor): Indicates atomic displacement or mobility (also not used by Qbics).
- Element Symbol: The chemical element symbol.
ATOM 1 N SER 10 9.630 40.740 60.970 0.00 0.00 N
ATOM 2 HT1 SER 10 9.940 40.000 61.630 0.00 0.00 H
ATOM 3 HT2 SER 10 8.910 41.340 61.420 0.00 0.00 H
ATOM 4 HT3 SER 10 10.450 41.360 60.880 0.00 0.00 H
ATOM 5 CA SER 10 9.180 40.240 59.600 0.00 0.00 C
ATOM 6 HA SER 10 9.510 39.210 59.580 0.00 0.00 H
Some software generates PDB files without the "Element Symbol" column. In such cases, Qbics will attempt to infer the element symbol from the "Atom Name" column.
However, this approach is risky and should be avoided, as "Atom Name" is, in principle, arbitrary and can be misleading. For example, CLA may refer to either a chlorine or a carbon atom, depending on the context of the PDB file.
Therefore, it is strongly recommended to always include the "Element Symbol" column in PDB files used for Qbics calculations.
Input Examples
Example: Transition State Search Using mol and mol2 to Specify Reactant and Product
Perform a transition state search using the NEB method at the xTB level of theory, with the reactant and product structures specified in mol and mol2, respectively:
opt
type neb
num_images 10
neb_k 0.01
end
mol # N-C-H
C 0. 0. 0.289
N 0. 0. -1.128
H 0. 0. 1.099
end
mol2 # H-N-C
C 0.403 -0.780 -0.690
N -0.021 0.041 0.036
H -0.382 0.739 0.654
end
xtb
chrg 0
end
task
opt xtb
endIn this file, the structures defined by mol and mol2 are shown below: