Tip
All input and output files can be downloaded here.
Density Functional Theory Calculations
This tutorial will lead you step by step to do quantum chemical calculations using Qbics.
The First Example: Water
We will now do the first calculation using Qbics. The following input file will do B3LYP-D3BJ/def2-tzvp energy for a water molecule:
# This first calculation.
basis
def2-tzvp
end
scf
charge 0
spin2p1 1
type R # You do not have to write it. The program will determine it by itself.
end
grimmedisp
type bj
end
mol
O 0. 0.00000000 -0.11081188
H 0. -0.78397589 0.44324751
H 0. 0.78397589 0.44324751
end
task
energy b3lyp
end
Then run it:
$ qbics water-1.inp -n 4 > water-1.out
Here, -n 4 means Qbics will use 4 CPU cores for parallization. This calculation is very fast. After calculation, you will find 2 new files:
water-1.out water-1.mwfn
water-1.out is the output file for this calculation. You can find energies, molecular orbital (MO) information, spin, population, electric multipole moments in it:
SCF Energies
============
Kinetic energy: 76.16685643 Hartree
Electron-nuclear attraction energy: -199.22128800 Hartree
Pseudopotential energy: 0.00000000 Hartree
Exchange-correlation energy: -7.55662337 Hartree
Electron Coulomb energy: 46.77766964 Hartree
Electron exchange energy: -1.78638759 Hartree
Nuclear repulsion energy: 9.15711600 Hartree
Grimme dispersion energy: -0.00057358 Hartree
----------------------------------------------------------------
SCF energy (E): -76.46323045 Hartree
Virial quotien (V/T): -2.00389112
Molecular Orbitals
==================
k = Gamma
HOMO-LUMO (5-6) gap: 8.990 eV
# Occupancies Energies/Hartree
1 2.000 -19.12516259
2 2.000 -1.01223986
3 2.000 -0.54372159
4 2.000 -0.38293836
...
Spin
====
Expected <S^2>: 0.00000; S = 0.00000
Calculated <S^2>: 0.00000; S = 0.00000
Mulliken Populations
====================
# Symbol Charge Spin
----------------------------------------------
1 O -0.64331377 0.00000000
2 H 0.32165689 0.00000000
3 H 0.32165689 0.00000000
----------------------------------------------
Sum -0.00000000 0.00000000
----------------------------------------------
Electric Multipole Moments
==========================
# Total Electronic Nuclear Unit
------------------------------------------------------------------------------------
Charge:
0 -0.0000 -10.0000 10.0000 |e|
Dipole moment:
X -0.0000 -0.0000 0.0000 Debye
Y 0.0000 0.0000 0.0000 Debye
Z 1.9936 1.9936 -0.0000 Debye
Totla 1.9936 Debye
Quadrupole moment:
XX -7.5861 -7.5861 0.0000 Debye*Angstrom
XY -0.0000 -0.0000 0.0000 Debye*Angstrom
XZ -0.0000 -0.0000 0.0000 Debye*Angstrom
YY -4.1639 -10.0682 5.9043 Debye*Angstrom
YZ 0.0000 0.0000 -0.0000 Debye*Angstrom
ZZ -6.4570 -8.8162 2.3592 Debye*Angstrom
------------------------------------------------------------------------------------
water-1.mwfn is the wave function file, which can be read by Qbics and Multiwfn. You can do all wave function visualization and analysis with Multiwfn and this file. For example, the following code will show MOs:
$ Multiwfn water-1.mwfn
$ 0
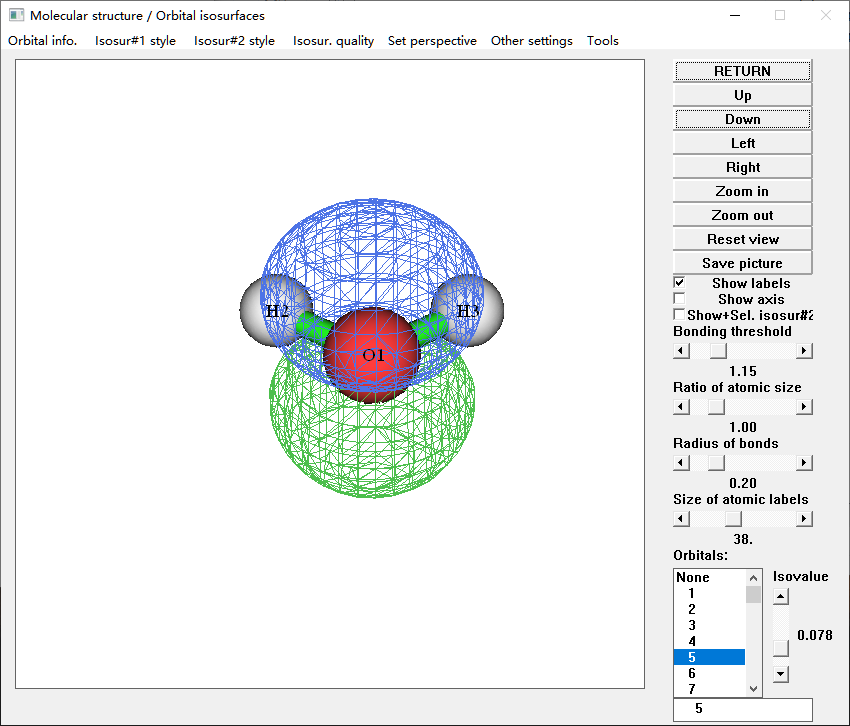
Explanation of The First Example
Now we will explain the input water-1.inp of the last example.
Anything after
#is treated as comments. You can write anything anywhere in the input file.basis ... endThis block define the basis set used the calculation. You can find all available basis sets inqbics/basis. Just input the file name. You can also define your own basis set, or set basis set for different elements. See below.scf ... endThis block controls how the self-consistent field (SCF) calculations are done. By its name, you may understand that:
chargeThe charge of the molecule, like0,+3,-1, etc.
spin2p1The spin multiplicity. For a molecule with \(n\) unpaired electrons, this total spin is \(S=\frac{n}{2}\), so spin multiplicity is \(2S+1 = n+1\). For example, for a triplet state of water, you will usespin2p1 3, then 2 unpaired electrons will occupy 2 alpha orbitals.
typeThe value isRif a restricted Hartree–Fock (HF) or Kohn–Sham (KS) is needed, orUis an unrestricted one is needed. If you do not write this, the program will determine it automatically according to spin multiplicity:Rfor singlet andUfor other cases
grimmedisp ... endThis block controls how Grimme dispersion correction is applied. Here we usebjto indicate Becke–Johnson dampling D3 correction.mol ... endThis block gives the molecule coordinates in XYZ format. You can also simply give a coordinate file name in XYZ or PDB format, saywater.xyzorwater.pdb.task ... endThis tells Qbics what it should do.energy b3lypmeans it will use B3LYP to calculate energy. You can put several tasks in this block.
Based on the explanations above, you can now try to calculate the triplet state of water:
# This first calculation.
basis
def2-tzvp
end
scf
charge 0
spin2p1 3
type U # If you do not write it. The program will determine it by itself.
end
grimmedisp
type bj
end
mol
O 0. 0.00000000 -0.11081188
H 0. -0.78397589 0.44324751
H 0. 0.78397589 0.44324751
end
task
energy b3lyp
end
You can find MO occupations and spin here:
Molecular Orbitals
==================
k = Gamma
Alpha HOMO-LUMO (6-7) gap: 3.247 eV
Beta HOMO-LUMO (4-5) gap: 5.963 eV
Alpha Alpha Beta Beta
# Occupancies Energies/Hartree Occupancies Energies/Hartree
1 1.000 -19.35044069 1.000 -19.31689276
2 1.000 -1.21472533 1.000 -1.12845421
3 1.000 -0.69876887 1.000 -0.66807740
4 1.000 -0.57036857 1.000 -0.52597568
5 1.000 -0.56380445 0.000 -0.30683178
6 1.000 -0.10311895 0.000 -0.01035402
...
Spin
====
Expected <S^2>: 2.00000; S = 1.00000
Calculated <S^2>: 2.00188; S = 1.00063
The spin of this calculation is 1.00063, which is very close to the theoretical value 1, indicating a very small spin contamination.
More Basis Set Configurations
Qbics hase provided flexible basis set configuration. This section will show it.
Define Your Own Basis Set
Assume you want to use a basis set called pc-2 for water. However, this is not available in qbics/basis, you can then define it by your self.
Go to basis set exchange website: https://www.basissetexchange.org/, choose
pc-2for H and O, and download it using GAUSSIAN format.
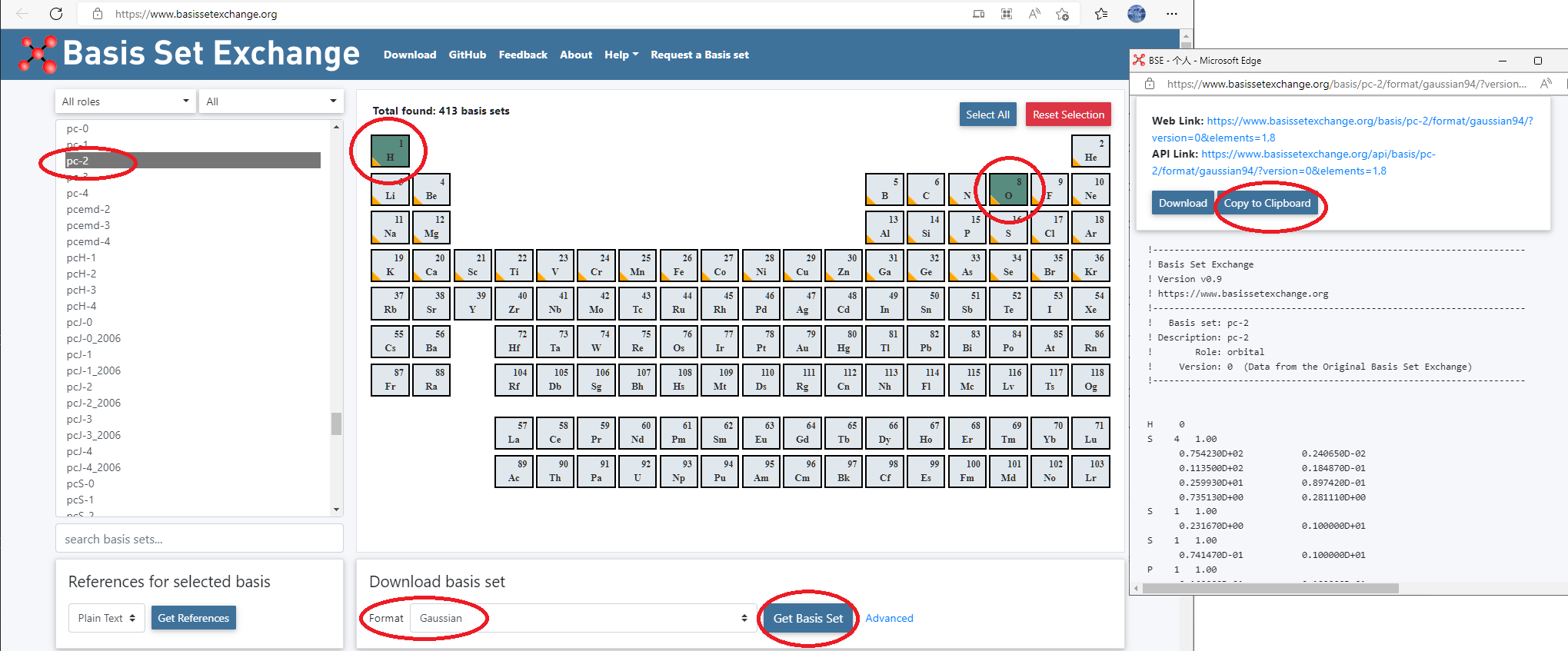
Attention
Replace all D to E in the basis set definitions!
Now we have two ways:
The first way: Simply copy it to
basis ... endblock:
# This first calculation.
basis
# From: https://www.basissetexchange.org/basis/pc-2/format/gaussian94/?version=0& elements=1,8
H 0
S 4 1.00
0.754230E+02 0.240650E-02
0.113500E+02 0.184870E-01
0.259930E+01 0.897420E-01
0.735130E+00 0.281110E+00
S 1 1.00
0.231670E+00 0.100000E+01
S 1 1.00
0.741470E-01 0.100000E+01
P 1 1.00
0.160000E+01 0.100000E+01
P 1 1.00
0.450000E+00 0.100000E+01
D 1 1.00
0.125000E+01 0.100000E+01
****
O 0
S 7 1.00
0.147820E+05 0.535190E-03
0.221730E+04 0.413750E-02
0.504740E+03 0.212450E-01
0.142870E+03 0.824530E-01
0.463000E+02 0.236710E+00
0.163370E+02 0.440390E+00
0.598280E+01 0.364650E+00
S 7 1.00
0.221730E+04 -0.192750E-05
0.504740E+03 -0.579640E-04
0.142870E+03 -0.794940E-03
0.463000E+02 -0.731250E-02
0.163370E+02 -0.405740E-01
0.598280E+01 -0.915940E-01
0.167180E+01 0.209400E+00
S 1 1.00
0.646620E+00 0.100000E+01
S 1 1.00
0.216690E+00 0.100000E+01
P 4 1.00
0.604240E+02 0.689490E-02
0.139350E+02 0.490050E-01
0.415310E+01 0.182550E+00
0.141580E+01 0.376330E+00
P 1 1.00
0.475490E+00 0.100000E+01
P 1 1.00
0.145290E+00 0.100000E+01
D 1 1.00
0.220000E+01 0.100000E+01
D 1 1.00
0.650000E+00 0.100000E+01
F 1 1.00
0.110000E+01 0.100000E+01
****
end
scf
charge 0
spin2p1 1
type R # You need not to write it. The program will determine it by itself.
end
grimmedisp
type bj
end
mol
O 0. 0.00000000 -0.11081188
H 0. -0.78397589 0.44324751
H 0. 0.78397589 0.44324751
end
task
energy b3lyp
end
Then do the calculation as usual.
The second way: Save the basis set in a file called, say,
pc-2-OH.txt. Assume you put it in/home/you/calc/pc-2-OH.txt, then write this file name inbasis ... endblock:
# This first calculation.
basis
/home/you/calc/pc-2-OH.txt
end
scf
charge 0
spin2p1 1
type R # You need not to write it. The program will determine it by itself.
end
grimmedisp
type bj
end
mol
O 0. 0.00000000 -0.11081188
H 0. -0.78397589 0.44324751
H 0. 0.78397589 0.44324751
end
task
energy b3lyp
end
Both ways will give the same result.
Different Basis Sets for Different Elements
Assume you want to use pc-2 (defined above) for oxygen but cc-pvdz for hydrogen, you can use the following basis ... end:
basis
element # This indicates that Qbics will assign basis set element by element.
O /home/you/calc/pc-2-OH.txt
H cc-pvdz
end
The word element in the first line of basis ... end block means that Qbics will assign basis set element by element. Then simply write the basis set for every element.
Use Pseudopotential
General Rules
For heavy elements, one should use pseudopotential for reasonable calculations.
Attention
You must write BOTH valence basis set and pseudopotential in the input file. If pseudopotential is not written explicitly in the input file, Qbics will NOT use it.
This is because for some elements, there exist both all-electron basis set and valence-only basis set with pseudopotential at the same time, so you should make it clear in the input file.
The valence basis sets and core pseudopotentials are stored in qbics/basis and qbics/pseudopotential, respectively. For an element, you must use them consistently:
Valence Basis Set |
Pseudopotential |
|---|---|
def2-X |
def2-ecp |
(aug-)cc-X-pp |
cc-ecp |
lanlX |
lanl-ecp |
For example, you can use def2-TZVPP and def-ecp for any element, but NEVER use def2-TZVPP and lanl-ecp together!
Assume you want to calculate B3LYP-D3BJ energy for \(AuCl_3\), you can use def2- series:
basis
def2-tzvp
end
pseudopotential
def2-ecp
end
scf
charge 0
spin2p1 1
end
grimmedisp
type bj
end
mol
Au 0.00000000 0.00000000 0.
Cl 0.00000000 -2.33000000 0.
Cl 2.01783919 1.16500000 0.
Cl -2.01783919 1.16500000 0.
end
task
energy b3lyp
end
Or, you may want to use cc-pvdz for chlorine and cc-pvdz-pp for gold, then the input is:
basis
element
Cl cc-pvdz
Au cc-pvdz-pp
end
pseudopotential
cc-ecp
end
This means that for the valence and core part of gold, cc-pvdz basis set and pseudopotential is used, respectively.
Define Your Own Pseudopotential
You can also define your own pseudopotential like we have done for basis set. Again for \(AuCl_3\), we want to use SBKJC valence basis set and pseudopotentials, then
Go to basis set exchange website: https://www.basissetexchange.org/, choose
SBKJC-VDZfor Au and Cl, and download it using GAUSSIAN format.
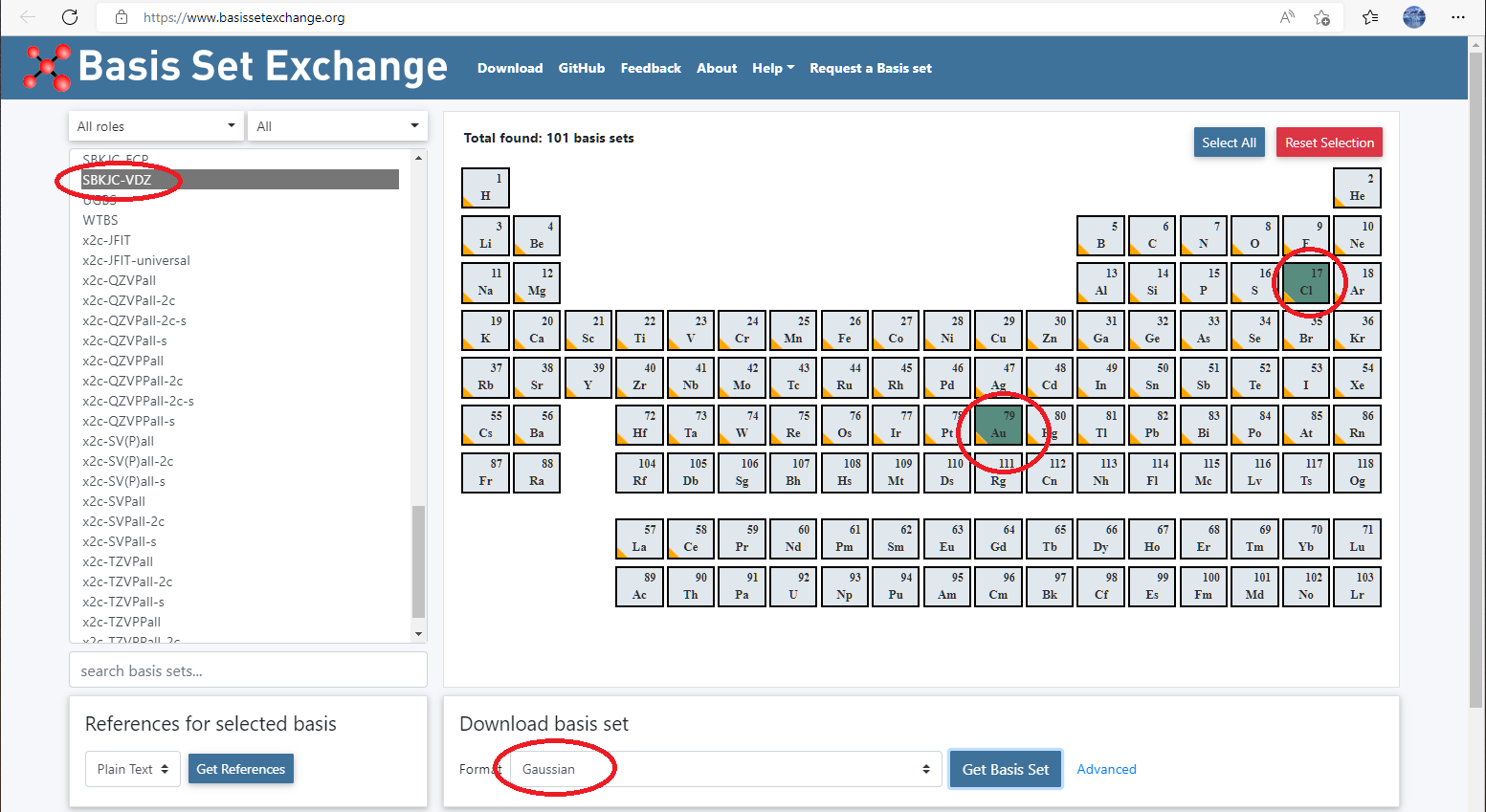
Paste basis set and pseudotential definitions in
basis ... endandpseudopotential ... endblock, respectively:
basis
# From: https://www.basissetexchange.org/basis/sbkjc-vdz/format/gaussian94/?version=0&elements=17,79
Cl 0
SP 3 1.00
2.225000 -.330980 -.126040
1.173000 0.115280 0.299520
0.385100 0.847170 0.583570
SP 1 1.00
0.130100 0.265340 0.340970
****
Au 0
SP 1 1.00
1.502000 1.000000 1.000000
SP 4 1.00
7.419000 0.222546 0.019924
4.023000 -1.086045 -.299997
1.698000 1.156039 0.748919
0.627100 0.518061 0.504023
SP 1 1.00
0.151500 1.000000 1.000000
SP 1 1.00
0.049250 1.000000 1.000000
D 3 1.00
3.630000 -.087402
1.912000 0.468634
0.842300 0.654805
D 1 1.00
0.375600 1.000000
D 1 1.00
0.154400 1.000000
****
end
pseudopotential
# From: https://www.basissetexchange.org/basis/sbkjc-vdz/format/gaussian94/?version=0&elements=17,79
CL 0
CL-ECP 2 10
d potential
1
1 4.8748300 -3.4073800
s-d potential
2
0 17.0036700 6.5096600
2 4.1038000 42.2778500
p-d potential
2
0 8.9002900 3.4286000
2 3.5264800 22.1525600
****
AU 0
AU-ECP 4 60
g potential
1
1 4.3876300 -10.7235800
s-g potential
3
0 1.5563600 6.3561200
2 3.7159300 -364.4403900
2 4.0679200 428.1975300
p-g potential
3
0 1.1879800 4.4151800
2 3.0155100 -136.5755000
2 3.5958800 194.2053500
d-g potential
2
0 35.2500000 8.8819800
2 5.0230700 86.7661200
f-g potential
1
0 1.6888100 6.2160300
****
end
scf
charge 0
spin2p1 1
end
grimmedisp
type bj
end
mol
Au 0.00000000 0.00000000 0.
Cl 0.00000000 -2.33000000 0.
Cl 2.01783919 1.16500000 0.
Cl -2.01783919 1.16500000 0.
end
task
energy b3lyp
end
Note, you MUST add **** between pseudopential definitions for each element.
Or, you can save both definitions in
/home/you/calc/SBJKC.txtand/home/you/calc/SBJKC-ECP.txt, respectively, and write these file names in the input file:
basis
/home/you/calc/SBJKC.txt
end
pseudopotential
/home/you/calc/SBJKC-ECP.txt
end
Good SCF Initial Guess
Qbics supports several ways to set SCF initial guess. The default one is the “superposition of atomic densities”, which works quiet well in most cases. Sometimes, one may or must use other ways.
Guess From Another Calculations
Consider \(CH_{3}NH_{3}^{+}OH^{-}\), we do a B3LYP-D3BJ/6-31g(d) calculation:
basis
6-31g(d)
end
scf
charge 0
spin2p1 1
end
grimmedisp
type bj
end
mol
C -0.02909313 -0.38166253 -1.63963825
H 0.82173570 -1.03039025 -1.62769450
H -0.03691367 0.17958863 -2.55059158
H -0.92347728 -0.96516294 -1.57252437
N 0.04484969 0.58404262 -0.44233044
H 0.93923384 1.16754303 -0.50944432
H -0.80597913 1.23277035 -0.45427419
H 0.05267023 0.02279146 0.46862289
O 0.06914388 -0.26694332 2.08310089
H -0.31212699 -0.12725829 2.95299779
end
task
energy b3lyp
end
After calculation, we will obtain a file ch3nh3oh-1.mwfn. We can use this as an initial guess for a more expensive calculation, like B3LYP-D3BJ/6-311g(2df,2pd) calculation:
basis
6-311g(2df,2pd)
end
scf
charge 0
spin2p1 1
end
scfguess
type mwfn
file ch3nh3oh-1.mwfn
end
grimmedisp
type bj
end
mol
C -0.02909313 -0.38166253 -1.63963825
H 0.82173570 -1.03039025 -1.62769450
H -0.03691367 0.17958863 -2.55059158
H -0.92347728 -0.96516294 -1.57252437
N 0.04484969 0.58404262 -0.44233044
H 0.93923384 1.16754303 -0.50944432
H -0.80597913 1.23277035 -0.45427419
H 0.05267023 0.02279146 0.46862289
O 0.06914388 -0.26694332 2.08310089
H -0.31212699 -0.12725829 2.95299779
end
task
energy b3lyp
end
Here, scfguess ... end block controls the initial guess with type, which can be:
Option |
Meaning |
|---|---|
|
Diagonalization of core Hamiltonian. NOT recommended. |
|
Superposition of atomic densities. Default. |
|
Read a wave function from a mwfn file given by |
|
Superposition of molecular fragment densities. See below. |
Attention
For an input file x.inp, the program will rewrite x.mwfn during the calculation, so if you want to keep the x.mwfn, you can copy it to something like x-1.mwfn, and write
scfguess
type mwfn
file x-1.mwfn
end
Then the program will read x-1.mwfn for initial guess but not overwrite x.mwfn.
Guess From Fragment Superposition
Consider \(CH_{3}NH_{3}^{+}OH^{-}\), it seems to be logical that one can use the superposition of \(CH_{3}NH_{3}^{+}\) and \(OH^{-}\) as initial guess. This is very easy:
basis
6-311g(2df,2pd)
end
scf
charge 0
spin2p1 1
end
scfguess
type fragden
frag +1 1 1-8
frag -1 1 9 10
end
grimmedisp
type bj
end
mol
C -0.02909313 -0.38166253 -1.63963825
H 0.82173570 -1.03039025 -1.62769450
H -0.03691367 0.17958863 -2.55059158
H -0.92347728 -0.96516294 -1.57252437
N 0.04484969 0.58404262 -0.44233044
H 0.93923384 1.16754303 -0.50944432
H -0.80597913 1.23277035 -0.45427419
H 0.05267023 0.02279146 0.46862289
O 0.06914388 -0.26694332 2.08310089
H -0.31212699 -0.12725829 2.95299779
end
task
energy b3lyp
end
Here, type fragden means we will use superposition of fragments as initial guess. The fragments can be defined with frag:
frag +1 1 1-8
The first and second number is the charge and spin multiplicity of the fragment. Then is the atom indices for this fargment. It can be discontinuous like:
frag +1 1 1 2 5 8-12 15
In this case, the atom 1 2 5 8 9 10 11 12 15 will be a fragment. You can define any numbers of fragments.
SCF Convergence Condition
There are 3 options in scf ... end block to control the convergence condition:
scf
energy_cov 1.E-6
density_cov 1.E-6
max_it 100
end
Here, energy_cov and density_cov means that the SCF is thought to converge when the energy and density change is smaller than 1.E-6 and 1.E-6, respectively. max_it means the maximum number of SCF iterations. You can change these when you need.